Understanding ALK structural data
“Know thy self, know thy enemy. A thousand battles, a thousand victories”; a quote by the famous Chinese historical figure Sun Tzu. Accordingly, scientists have been working hard at understanding the fundamentals of the ALK protein.
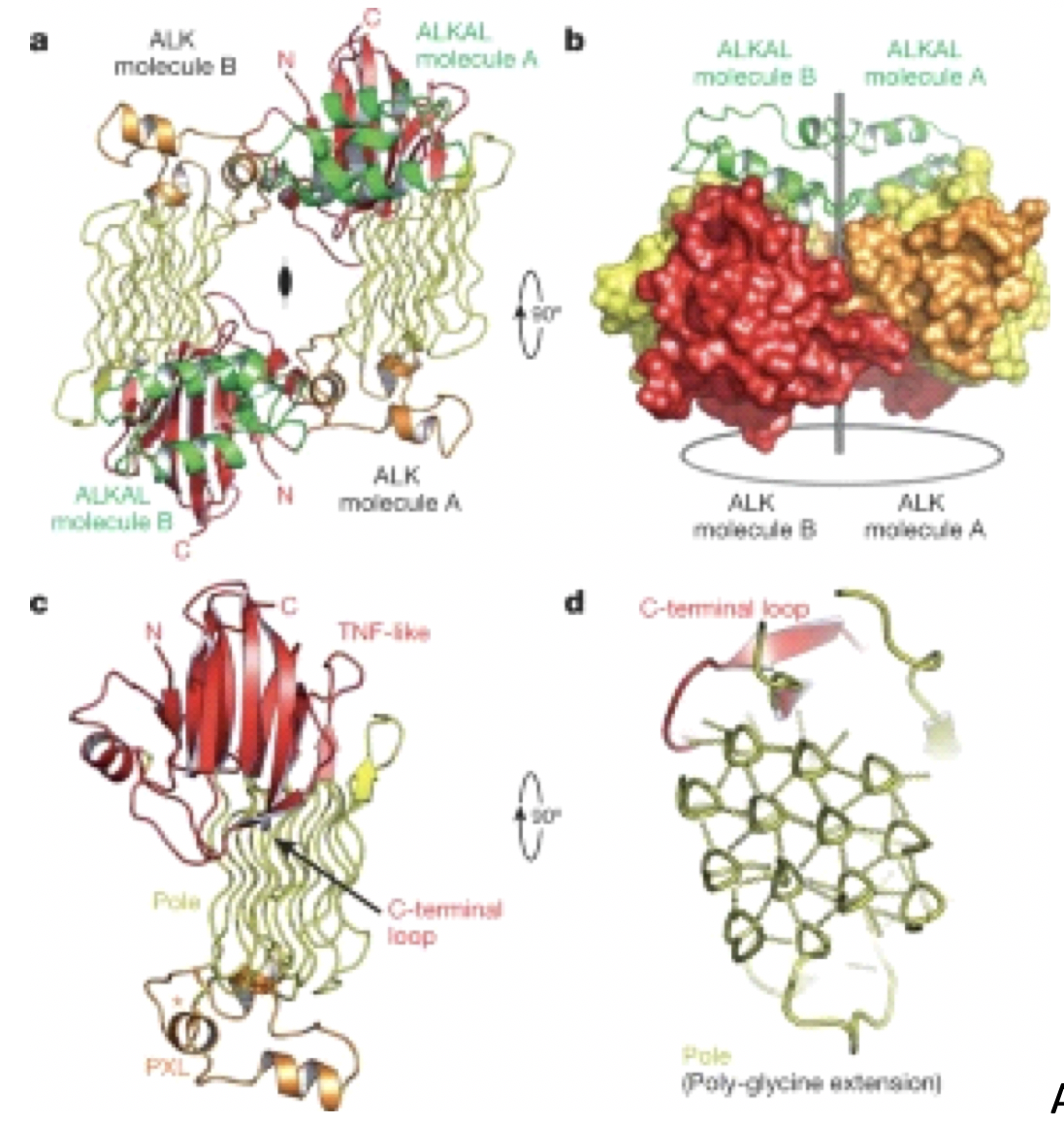
Recently, a paper was published by a group of scientists in Belgium lead by Dr. Savvides, in which they described the newly discovered 3D structure of the activated ALK complex. The binding of ALK and its ligand forms a dimeric assembly with a folded symmetry that can “tent” a specific cytokine molecule to the cell surface. This basic understanding of how the protein binds and behaves can help scientists make medicines that are more specific to ALK.
A group from Yale also recently published additional structural information on the various domains of ALK and how the ligand and receptor interact. They found a terminal region termed polyglycine extension loop (PXL) which is important in ALK dimerization. If you can abolish dimerization, they hypothesize that ALK activation can be eliminated.
In neuroblastoma, the extracellular domain of ALK was found to be cut off, and it resulted in downregulation of nuclear β-catenin, and the Endothelial-Mesenchymal Transition (EMT) genes to be altered. Thus, cells are more likely to migrate. This location is specific to Asn654-Leu655.
This basic understanding of how the ALK protein binds and behaves will help scientists create better treatments in the future.
https://www.nature.com/articles/s41586-021-04141-7
https://www.sciencedirect.com/science/article/pii/S2211124721007610
https://www.nature.com/articles/s41586-021-03959-5
Author: Alice Chou